GenExpTimecourses: Analysis of gene expression time-courses
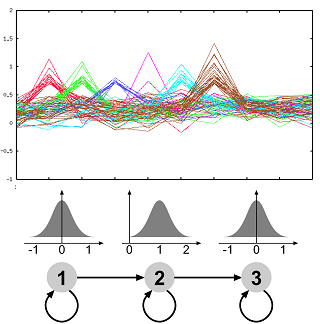 The molecular processes of life are dynamic over time. Microarray experiments measuring the expression levels of a multitude of genes over time are one way of gaining insight into the dynamic processes. As a first analysis groups of similar expression patterns are routinely identified. We have developed an approach which allows to use prior knowledge, is flexible and very robust to noise. The method is implemented in the software GQL which allows control of the analysis process by use of graphical user interfaces. Currently, we are extending our framework to allow integration of further data related to transcription or protein interactions. Furthermore, we are also investigating methodologies for validating clustering of genes with functional annotation.
The molecular processes of life are dynamic over time. Microarray experiments measuring the expression levels of a multitude of genes over time are one way of gaining insight into the dynamic processes. As a first analysis groups of similar expression patterns are routinely identified. We have developed an approach which allows to use prior knowledge, is flexible and very robust to noise. The method is implemented in the software GQL which allows control of the analysis process by use of graphical user interfaces. Currently, we are extending our framework to allow integration of further data related to transcription or protein interactions. Furthermore, we are also investigating methodologies for validating clustering of genes with functional annotation.
For further information contact Alexander Schliep (alexander@schlieplab.org). This project is connected to the following projects: GQL, GHMM, pGQL.
Team
Members: Alexander Schliep, Ivan G Costa, Christoph Hafemeister. Collaborators: Alexander Schönhuth (Centrum Wiskunde & Informatica).
Publications
Costa et al.. Constrained Mixture Estimation for Analysis and Robust Classification of Clinical Time Series. Bioinformatics 2009, 12:25, i6–14. (ISMB 2009). Schönhuth et al.. Semi-supervised Clustering of Yeast Gene Expression. In Cooperation in Classification and Data Analysis, Springer, 151–160, 2009. Proceedings of Two German-Japanese Workshops . Costa. Mixture Models for the Analysis of Gene Expression: Integration of Multiple Experiments and Cluster Validation. Ph.D. Thesis, Freie Universität Berlin, May 2008. Costa et al.. On the feasibility of Heterogeneous Analysis of Large Scale Biological Data. In Proceedings of ECML/PKDD 2006 Workshop on Data and Text Mining for Integrative Biology, 55–60, 2006. Costa et al.. The Graphical Query Language: a tool for analysis of gene expression time-courses. Bioinformatics 2005, 21:10, 2544–5. Schliep et al.. Analyzing gene expression time-courses. IEEE/ACM Trans Comput Biol Bioinform 2005, 2:3, 179–193. Schliep et al.. Robust inference of groups in gene expression time-courses using mixtures of HMMs. Bioinformatics 2004, 20 Suppl 1, i283–i289. Schliep et al.. Using hidden Markov models to analyze gene expression time course data. Bioinformatics 2003, 19 Suppl 1, i255–i263.