GHMM: General Hidden Markov Model library
The General Hidden Markov Model library (GHMM) is a freely available LGPL-ed C library implementing efficient data structures and algorithms for basic and extended HMMs. The development is hosted at Sourceforge http://sourceforge.net/projects/ghmm/, where you have access to the Subversion repository, mailing lists and forums.
Features
- Discrete and continous emissions
- Mixtures of PDFs for continous emissions
- Non-homogenous Markov chains
- Pair HMMs
- Clustering and mixture modelling for HMMs
- Graphical Editor HMMEd
- Python bindings
- XML-based file format
- Portable (autoconf, automake)
HMMEd (the Hidden Markov Model editor) is a graphical application which allows to create and edit Hidden Markov Models. Supported are
- discrete emissions on arbitrary alphabets; this includes DNA, RNA, and amino acids, and
- continous scalar emissions.
Discrete emission probabilities, as well as transition probabilities can be graphically edited using pie charts with handles. Parameters can also be entered directly. HMM topology can be edited manually by adding/deleting states and connecting them with the mouse. Hierarchical models, i.e. super nodes corresponding to complete promoters or codon models in gene finding HMMs, will be supported. Currently, a visually pleasant layout has to be done by hand.
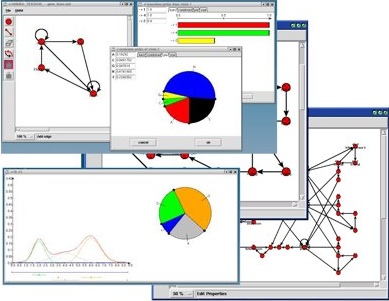
For models with continous emission, a graphical editor for mixtures of pdfs is under development. A screenshot from a working prototype is shown above. The handles underneath the pdfs change mean as well as other pdf parameters, the pie chart controls the mixture coefficients.
HMMEd is licensed under the GPL. Its based on code from Gato the graph animation toolbox. Note: HMMEd can be found in the Gato subdirectory in the CVS [Browse Sourceforge CVS]
For further information see the main website at http://ghmm.org/ or contact Alexander Schliep (alexander@schlieplab.org). This software is a result of or used in the following projects: GQL, ArrayCGH, GenExpTimecourses, GeneFinder, pGQL, HaMMLET.
Team
Members: Alexander Schliep, Alexander Schliep, Ivan G Costa, Wasinee Rungsarityotin, Benjamin Georgi, Janne Grunau, Christoph Hafemeister, Daniel Z Kaplan. Collaborators: Alexander Schönhuth (Centrum Wiskunde & Informatica).
Publications
Costa. Mixture Models for the Analysis of Gene Expression: Integration of Multiple Experiments and Cluster Validation. Ph.D. Thesis, Freie Universität Berlin, May 2008. Costa et al.. Semi-supervised learning for the identification of syn-expressed genes from fused microarray and in situ image data. BMC Bioinformatics 2007, 8, S3. Costa et al.. The Graphical Query Language: a tool for analysis of gene expression time-courses. Bioinformatics 2005, 21:10, 2544–5. Schliep et al.. Analyzing gene expression time-courses. IEEE/ACM Trans Comput Biol Bioinform 2005, 2:3, 179–193. Schliep et al.. The General Hidden Markov Model Library: Analyzing Systems with Unobservable States. In Forschung und wissenschaftliches Rechnen: Beiträge zum Heinz-Billing-Preis 2004, Gesellschaft für wissenschaftliche Datenverarbeitung, 121–135, 2005. Schliep et al.. Robust inference of groups in gene expression time-courses using mixtures of HMMs. Bioinformatics 2004, 20 Suppl 1, i283–i289. Riemer. Chromosome-wide Expression for Improving ab-initio Gene Prediction. Bachelor's Thesis, Freie Universität Berlin, 2004. Grunau. Discriminative Learning in Hidden Markov Models. Bachelor's Thesis, Freie Universität Berlin, 2004. Schliep et al.. Using hidden Markov models to analyze gene expression time course data. Bioinformatics 2003, 19 Suppl 1, i255–i263. Weisse. Recognition of Circular Permutations in Proteins with Hidden Markov Models. Bachelor's Thesis, Freie Universität Berlin, 2003. Georgi. A Graph-Based Approach to Clustering of Profile Hidden Markov Models. Bachelor's Thesis, Freie Universität Berlin, 2003. Knab et al.. Model-Based Clustering With Hidden Markov Models and its Application to Financial Time-Series Data. In Between Data Science and Applied Data Analysis, Springer, 561–569, 2003. Proceedings of the GfKl 2002. Wichern. Hidden Markov Models for the analysis of data from saving and loan banks. Ph.D. Thesis, University of Cologne, 2001. In German.. Knab. Extension of Hidden Markov Models for the analysis of financial time-series data. Ph.D. Thesis, University of Cologne, 2000. In German..