AlgoEngineering: Reduced representations and cache-efficient algorithms and data structures for bioinformatics
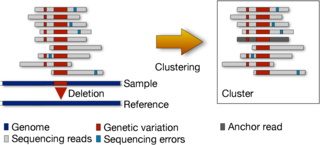 Reduced representations Analyzing large genomic datasets require enormous amount of computational resources in terms of running time, impact on cache, memory and disk space. We are working on finding alternate, reduced representations of these datasets which will enable downstream applications to work much more efficiently.
Reduced representations Analyzing large genomic datasets require enormous amount of computational resources in terms of running time, impact on cache, memory and disk space. We are working on finding alternate, reduced representations of these datasets which will enable downstream applications to work much more efficiently.
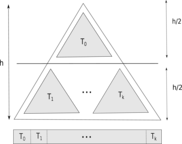 Cache efficient algorithms We are also investigating the effect of limited cache on bioinformatics tools, and looking for ways to overcome the difficulties it poses to k-mer counting and read mapping problem. Cache efficiency can play a very important role in designing computationally feasible tools for handling large datasets. With Turtle, we demonstrated that asymptotically more expensive algorithms can outperform less expensive algorithms by being cache efficient.
Cache efficient algorithms We are also investigating the effect of limited cache on bioinformatics tools, and looking for ways to overcome the difficulties it poses to k-mer counting and read mapping problem. Cache efficiency can play a very important role in designing computationally feasible tools for handling large datasets. With Turtle, we demonstrated that asymptotically more expensive algorithms can outperform less expensive algorithms by being cache efficient.
For further information contact Alexander Schliep (alexander@schlieplab.org). This project is connected to the following projects: BayesianHMM, TreQ, SLiQ, HTSMethods, Turtle, TreqCG.
Team
Members: Alexander Schliep, Md P. Mahmud, Rajat S Roy, John Wiedenhoeft.
Publications
Mahmud et al.. TreQ-CG: Clustering Accelerates High-Throughput Sequencing Read Mapping. Technical report, 2014. Mahmud et al.. Speeding Up Bayesian HMM by the Four Russians Method. In Algorithms in Bioinformatics, Springer Berlin / Heidelberg, 6833, 188–200, 2011. Mahmud et al.. Fast MCMC Sampling for Hidden Markov Models to Determine Copy Number Variations. BMC Bioinformatics 2011, 12:1, 428.