HTSMethods: Analysis of high-throughput sequencing data
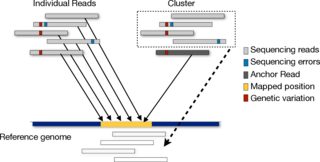 Next generation sequencing experiments produce millions of short reads from target genomes in a cost-efficient manner. Higher throughput brings new challenges such as how to map these short reads efficiently and how to deal with errors introduced by sequencing machines. Currently, we are investigating the problem of read-mapping in an indel-tolerant manner. We are also looking for techniques which will efficiently map reads back to reference genome.
Next generation sequencing experiments produce millions of short reads from target genomes in a cost-efficient manner. Higher throughput brings new challenges such as how to map these short reads efficiently and how to deal with errors introduced by sequencing machines. Currently, we are investigating the problem of read-mapping in an indel-tolerant manner. We are also looking for techniques which will efficiently map reads back to reference genome.
For further information contact Md P. Mahmud (pavelm@cs.rutgers.edu). This project is connected to the following projects: TreQ, SLiQ, Turtle, TreqCG, AlgoEngineering.
Team
Members: Md P. Mahmud, John Wiedenhoeft. Collaborators: Alexander Schönhuth (Centrum Wiskunde & Informatica), Gunnar Klau (Centrum Wiskunde & Informatica), Debashish Bhattacharya (Department of Ecology, Evolution, and Natural Resources, Rutgers), Shridar Ganesan (The Cancer Institute of New Jersey), Kevin Chen (Department of Genetics, Rutgers).
Publications
Damaschke et al.. An Optimization Problem Related to Bloom Filters With Bit Patterns. In SOFSEM 2018: Theory and Practice of Computer Science, Springer, 10706, 525–538, Jan 2018. Mahmud et al.. TreQ-CG: Clustering Accelerates High-Throughput Sequencing Read Mapping. Technical report, 2014. Roy et al.. Turtle: Identifying frequent k-mers with cache-efficient algorithms. Bioinformatics 2014, 14:30, 1950–7. Roy et al.. Turtle: Identifying frequent k-mers with cache-efficient algorithms.. Technical report, May 2013. Arxiv. Marshall et al.. CLEVER: Clique-Enumerating Variant Finder. Bioinformatics 2012, 28:22, 2875–82.. Marshall et al.. CLEVER: Clique-Enumerating Variant Finder. Technical report, Jul 2012. Arxiv. Roy et al.. SLIQ: Simple Linear Inequalities for Efficient Contig Scaffolding. Journal of Computational Biology 2012, 19, 1162–75. Mahmud et al.. Indel-tolerant Read Mapping with Trinucleotide Frequencies using Cache-Oblivious kd-Trees. Bioinformatics 2012, 28:18, i325–i332.