The SchliepLAB is part of the Faculty of Health Sciences Brandenburg and located at the Brandenburg University of Technology Cottbus-Senftenberg. Part of the group is at the department for Computer Science and Engineering, which is a joint department of the University of Gothenburg and Chalmers University of Technology.
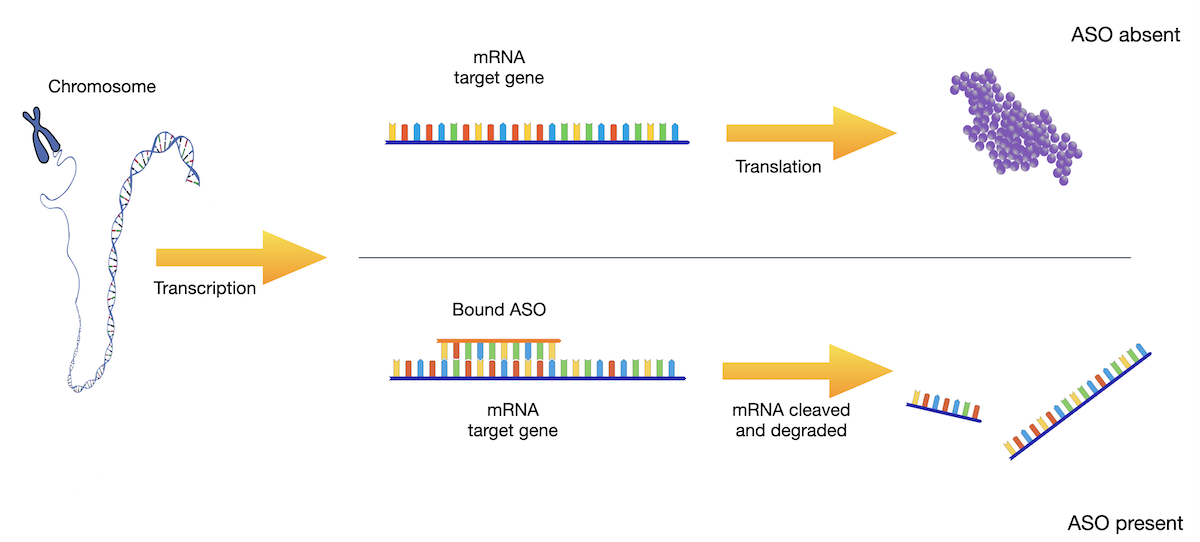
AI for design of oligonucleotide therapeutics
In particular for Antisense-Oligonucleotides (ASO) acting via RNAse H1-mediated knockdown binding energies and kinetics of ASO-mRNA duplexes are crucial for predicting efficacy and safety. We predict binding energies from sequence, study kinetics of ASO action to make the design process of ASO drugs more predictable, and in collaborative projects combine molecular dynamics and AI to extend predictive models for a wider range of nucleotide modifications. Our federated, privacy-preserving learning approach allows competitors to pool data in the training of predictors for binding energies.
ML for pan-genome graphs
Pan-genome graphs provide a principled approach to dealing with structural variants and the high degree of diversity between genomes. ML on pan-genome graphs will allow to tackle prediction and regression tasks for diverse populations including quantities relevant for oligonucleotide therapeutics such as transcription speed or accessibility as well as clinically relevant variables.
ML and Algorithmics for Sequencing Data
The data created from high-throughput experimental platforms such as high-throughput sequencing (HTS) poses computational challenges in particular if advanced statistical approaches such as Bayesian methods are used in analysis. Previously we developed a compressive genomics approach funded by the Big Data to Knowledge Program (BD2K) of the NIH, used Wavelet-compression in Bayesian HMMs for detecting copy number variants, and greatly improved the utility of statistical ML models representing genomes—variable length Markov chains—by faster algorithms for learning. This allows for example alignment-free genome comparisons from raw reads.
Education
Computational thinking is a core requirement across disciplines. Teaching computational and algorithmic ideas can benefit greatly from software tools. We develop animation systems for graph algorithms which are available on the desktop, as a web app, and soon as an iOS App; CATBox is a Springer textbook using Gato. Learners can concentrate on tackling exciting bioinformatics problems with our Hidden Markov Model library.
Further information
Find out about the people in the lab, our research areas, our publications, the software tools we develop and maintain or how to contact us.
Presentations:
- Sept. 24, 2024 at WASP Gothenburg, Sweden
- Sept. 24, 2024 at WASP Gothenburg, Sweden
- Feb. 22, 2024 at Zentrum für Künstliche …
- March 6, 2020 at Department of Computer …
- Nov. 28, 2019 at Practice of Enterprise …
- Jan. 9, 2019 at Conference on Teaching …
- more ...
Recent publications:
- Predicting oligonucleotide thermodynamics with discrete …
- Multiplexed ASOs with complementary off-targets
- de Novo Generated Combinatorial Library …
- Evolution shapes and conserves genomic …
- Compressed computations using wavelets for …
- AI for infectious diseases: Deep …
- more ...
Upcoming events:
- Bioinformatics 2026, Mar. 2-4, 2026, Marbella, Spain
- ISBCB 2026, Mar. 26-29, 2026, Kitakyushu, Japan
- ISCB UK, Apr. 21-22, 2026, Cambridge, UK
- RECOMB 2026, May 26-29, 2026, Thessaloniki, Greece
- ACM-BCB 2026, June 30-July 3, 2026, University of Calabria, Rende (Cosenza), Italy
- ISMB 2026, July 12-16, 2026, Washington D.C., USA
- more ...