GQL: Graphical Query Language
GQL is a suite of tools for analyizing time-course experiments. Currently, it is adapted to gene expression data. The two main tools are GQLQuery, for querying data sets, and GQLCluster, which provides a way for computing groupings based on a number of methods (model-based clustering using HMMs as cluster models and estimation of a mixture of HMMs).
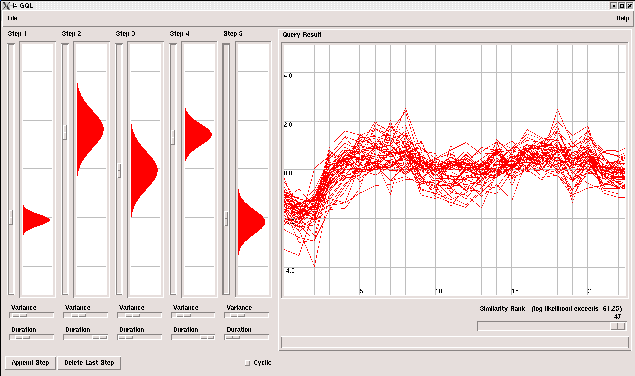
GQLQuery: Querying time-courses
The GUI has been ported to Python using Tkinter and the brand-new Python bindings for GHMM. It runs on all Linux/Unix boxes. Executable binaries for MAC and Windows are provided.
GQLCluster: Finding groups in time-courses

For further information see the main website at http://ghmm.org/gql or contact Ivan G Costa (filho@molgen.mpg.de). This software is a result of or used in the following projects: GenExpTimecourses, GHMM, pGQL.
Team
Members: Ivan G Costa, Alexander Schliep, Ivan G Costa, Ruben B. Schilling. Collaborators: Alexander Schönhuth (Centrum Wiskunde & Informatica).
Publications
Schönhuth et al.. Semi-supervised Clustering of Yeast Gene Expression. In Cooperation in Classification and Data Analysis, Springer, 151–160, 2009. Proceedings of Two German-Japanese Workshops . Costa. Mixture Models for the Analysis of Gene Expression: Integration of Multiple Experiments and Cluster Validation. Ph.D. Thesis, Freie Universität Berlin, May 2008. Costa et al.. Semi-supervised learning for the identification of syn-expressed genes from fused microarray and in situ image data. BMC Bioinformatics 2007, 8, S3. Costa et al.. The Graphical Query Language: a tool for analysis of gene expression time-courses. Bioinformatics 2005, 21:10, 2544–5. Schliep et al.. Analyzing gene expression time-courses. IEEE/ACM Trans Comput Biol Bioinform 2005, 2:3, 179–193. Schliep et al.. The General Hidden Markov Model Library: Analyzing Systems with Unobservable States. In Forschung und wissenschaftliches Rechnen: Beiträge zum Heinz-Billing-Preis 2004, Gesellschaft für wissenschaftliche Datenverarbeitung, 121–135, 2005. Schliep et al.. Robust inference of groups in gene expression time-courses using mixtures of HMMs. Bioinformatics 2004, 20 Suppl 1, i283–i289. Schliep et al.. Using hidden Markov models to analyze gene expression time course data. Bioinformatics 2003, 19 Suppl 1, i255–i263.