CSIMixtures: Context-specific independence mixture modeling for sequence motifs
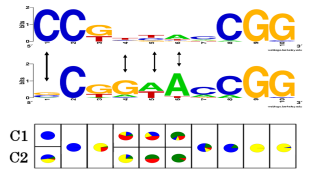 The modeling and analysis of sequence motives is one central task in the elucidation of biological processes such as gene regulation.
The choice of model class is crucial to obtain a representation of the motive suitable for the biological application. For instance previous studies showed that for transcription factors which bind to divergent binding sites, mixtures of multiple PWMs increase performance. However, estimating a conventional mixture distribution for each position will in many cases cause overfitting. We avoid this problem by employing a context-specific independence (CSI) framework. In CSI mixtures model complexity is automatically adapted to match the variability found in a given data set.
The modeling and analysis of sequence motives is one central task in the elucidation of biological processes such as gene regulation.
The choice of model class is crucial to obtain a representation of the motive suitable for the biological application. For instance previous studies showed that for transcription factors which bind to divergent binding sites, mixtures of multiple PWMs increase performance. However, estimating a conventional mixture distribution for each position will in many cases cause overfitting. We avoid this problem by employing a context-specific independence (CSI) framework. In CSI mixtures model complexity is automatically adapted to match the variability found in a given data set.
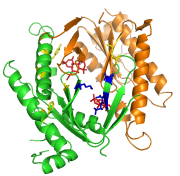 Another application of the CSI mixture framework is clustering of protein families for simultaneous inference of subgroups and prediction of specificity determining residues based on multiple sequence alignments of protein families. A Dirichlet mixture prior based on nine basic chemical properties of the standard amino acids is used to regularize the structure learning for protein domain data. Evaluation of the method on several well studied families revealed a good clustering performance and ample biological support for the predicted positions.
Another application of the CSI mixture framework is clustering of protein families for simultaneous inference of subgroups and prediction of specificity determining residues based on multiple sequence alignments of protein families. A Dirichlet mixture prior based on nine basic chemical properties of the standard amino acids is used to regularize the structure learning for protein domain data. Evaluation of the method on several well studied families revealed a good clustering performance and ample biological support for the predicted positions.
For further information contact Benjamin Georgi (georgi@molgen.mpg.de). This project is connected to the following projects: PyMix.
Team
Members: Benjamin Georgi, Alexander Schliep. Collaborators: Jörg Schultz (Universitaet Wuerzburg, Biozentrum), Pamela Flodman (UC Irvine, Human Genetics), M. Anne Spence (UC Irvine, Human Genetics).
Publications
Georgi et al.. Partially-supervised context-specific independence mixture modeling. In workshop on Data Mining in Functional Genomics and Proteomics, ECML 2007, 2007. Georgi et al.. Mixture model based group inference in fused genotype and phenotype data. In Studies in Classification, Data Analysis, and Knowledge Organization, Springer, 2007. Georgi et al.. Context-Specific Independence Mixture Modelling for Protein Families. In Knowledge Discovery in Databases: PKDD 2007, Springer Berlin / Heidelberg, Volume 4702/2007, 79–90, 2007. Georgi et al.. Context-specific independence mixture modeling for positional weight matrices. Bioinformatics 2006, 22:14, e166–e173.