CellDiff: Understanding transcriptional regulation in cell differentiation
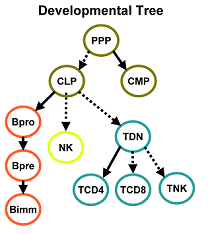 The regulatory processes that govern cell proliferation and differentiation are central to developmental biology. Particularly well studied in this respect is the lymphoid system due to its importance for basic biology and for clinical applications. Gene expression measured in lymphoid cells in several distinguishable developmental stages helps in the elucidation of underlying molecular processes, which change gradually over time and lock cells in either the B cell, T cell or Natural Killer cell lineages. Large-scale analysis of these gene expression trees requires computational support for tasks ranging from visualization, querying, and finding clusters of similar genes, to answering detailed questions about the functional roles of individual genes.
The regulatory processes that govern cell proliferation and differentiation are central to developmental biology. Particularly well studied in this respect is the lymphoid system due to its importance for basic biology and for clinical applications. Gene expression measured in lymphoid cells in several distinguishable developmental stages helps in the elucidation of underlying molecular processes, which change gradually over time and lock cells in either the B cell, T cell or Natural Killer cell lineages. Large-scale analysis of these gene expression trees requires computational support for tasks ranging from visualization, querying, and finding clusters of similar genes, to answering detailed questions about the functional roles of individual genes.
We present the first statistical framework designed to analyze gene expression data as it is collected in the course of lymphoid development through clusters of co-expressed genes and additional heterogeneous data. We introduce dependence trees for continuous variates, which model the inherent dependencies during the differentiation process naturally as gene expression trees. Such trees can have their structure estimated from the data or derived from expert knownledge. Several trees are combined in a mixture model to allow inference of potentially overlapping clusters of co-expressed genes. Computational results for several data sets from the lymphoid system demonstrate the relevance of this framework. We recover well-known biological facts and identify promising novel regulatory elements of genes and their functional assignments.
For further information contact Ivan G Costa (filho@molgen.mpg.de).
Team
Members: Ivan G Costa, Alexander Schliep, Ivan G Costa, Christoph Hafemeister. Collaborators: Fritz Melchers (Max Planck Institute for Infection Biology ).
Publications
Costa. Mixture Models for the Analysis of Gene Expression: Integration of Multiple Experiments and Cluster Validation. Ph.D. Thesis, Freie Universität Berlin, May 2008. Costa et al.. Gene expression trees in lymphoid development. BMC Immunol 2007, 8:1, 25. Hafemeister. Structure Learning of Conditional Trees. Bachelor's Thesis, Freie Universität Berlin, Jun 2006.